Our Research
Our lab works on three interrelated subjects: (1) DNA Repair Enzymology and Genomics; (2) Mammalian Circadian Clock; (3) Control of DNA Repair by the Circadian Clock.
DNA Repair Enzymology and Genomics
DNA damage by exogenous physical and chemical agents is the most common cause of cancer. Conversely, some of the most commonly used anticancer drugs kill malignant cells by damaging their DNA. DNA Repair is the ensemble of molecular mechanisms that eliminate DNA damage from the genome, and it plays crucial roles in carcinogenesis and in cancer therapy. Our lab works on Nucleotide Excision Repair which is the sole pathway for repairing cyclobutane pyrimidine dimers (CPDs) and cisplatin 1,2-d(GpG) adducts that cause cancer and cure cancer, respectively. We discovered that these lesions are removed from DNA by dual incisions that generate 24-32 nucleotide-long oligomers (“nominal 30-mer”). We identified and purified the 6 repair factors, RPA, XPA, XPC, TFIIH, XPG, XPF-ERCC1, that are essential for dual incisions. Using these purified factors, we reconstituted human excision repair in vitro and defined the molecular mechanism of excision repair. The in vitro work was complemented by in vivo studies which enabled us to generate a repair map of the entire genome. We discovered that the nominal 30-mer is released in a tight complex with TFIIH, and this finding enabled us to isolate the nominal 30-mers from irradiated cells and subject them to deep sequencing (Fig. 1). By using normal human fibroblasts and mutant cell lines we created repair maps for general repair and transcription-coupled repair of UV damage for the entire human genome through this method which we have named XR-seq (eXcision Repair-Sequencing). Our future work will exploit XR-seq to uncover novel genomic regulators of excision repair of DNA damage by anti-cancer drugs with the ultimate goal of developing improved chemotherapy regimens. In addition, we plan to investigate the effect of the excised nominal 30-mer on cellular physiology and the processing and ultimate fates of the excised oligonucleotides.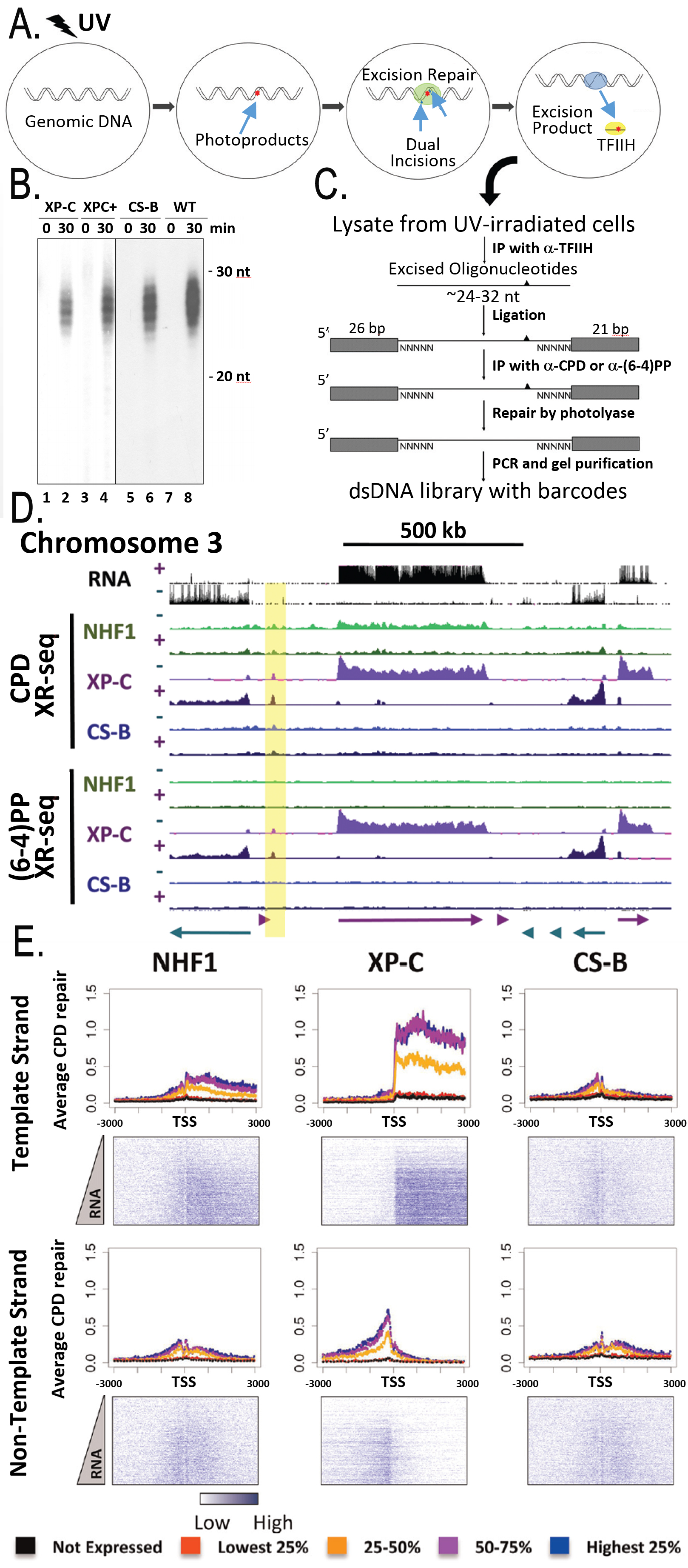
Fig. 1. The XR-seq method. (A) Schematic of nucleotide excision repair. (B) Excision patterns of photoproducts in wild-type, XP-C (deficient in global re-pair), and CS-B (deficient in TCR) cells. (C) Procedure for preparation of the dsDNA library for the Illumina HiSeq 2000 platform. (D) Distribution of the XR-seq signal, separated by strand, for CPD (top) and (6-4)PP (bot-tom) over a 1.5-Mb region of chromosome 3. (E) Strong association of TCR with RNA levels.
Mammalian Circadian Clock
The circadian clock is the internal timekeeping system that controls cyclic changes in physiology and behavior to prepare the organism for the unique challenges of the solar day. In mice and humans the circadian rhythm at the organismal level is generated by a molecular clock of periodicity of ~24 hrs. The molecular clock consists of a transcription-translation feedback loop (TTFL) in which the heterodimeric transcriptional activator CLOCK-BMAL1 promotes transcription of transcriptional repressors, CRY (Cryptochrome) and PER (Period) which counter the activity of CLOCK-BMAL1 as shown in Fig. 3A. Our group discovered that CRY is a core human clock protein and that it mediates repression by two mechanisms (Fig. 3B,C): In one CRY binds to the CLOCK-BMAL1 complex on DNA and blocks its interaction with the transcription machinery. In the second mode of repression, the co-repressor PER displaces the entire activator complex from the promoter in a CRY-dependent manner. Our research has provided a mechanistic basic for how this dual repression mechanism confers precision and resilience and, at the same time, flexibility and adaptability to the signaling pathways and networks and therefore influences physio-pathologic conditions ranging from sleep regulation and jetlag to metabolic syndrome to cancer. We discovered that the circadian clock regulates nucleotide excision repair and hence the susceptibility to UV-induced skin cancer as a function of time of the day of exposure to light. Because excision repair is also the main repair mechanism for removing DNA lesions from the genome that are generated by the anti-cancer drug cisplatin, we are currently working at translating our finding of clock-excision repair connection to develop improved chemotherapy regimens.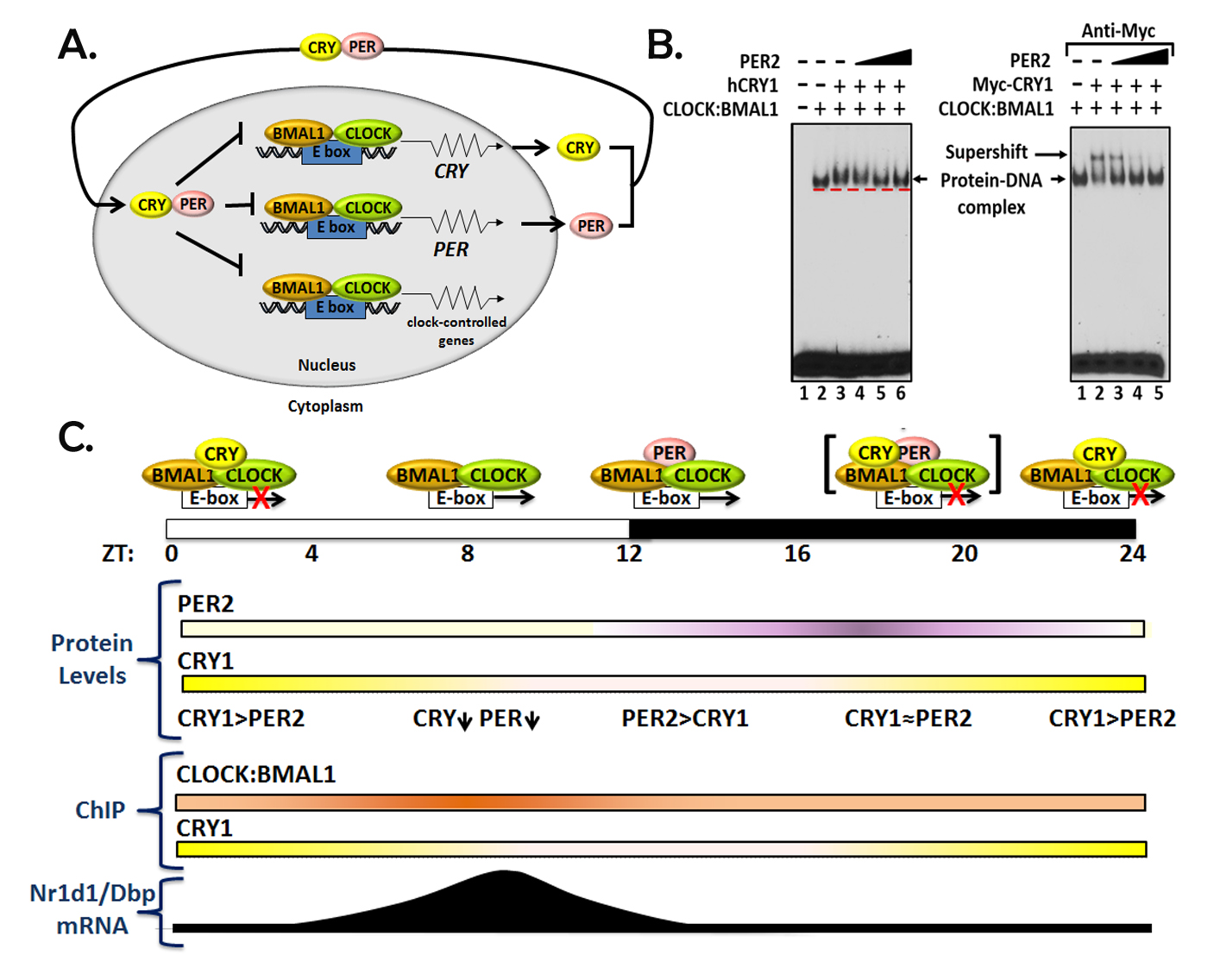
Fig. 2. Current Model for the Mammalian Circadian Clock. (A) Mammalian molecular clock model of the transcription-translation feedback loop (TTFL). (B) PER2 removes CRY1 from the CRY1:CLOCK:BMAL1:E-box complex. (C) New model for the mammalian circadian clock. The figure shows a semiquantitative heat map representation of CRY1 and PER2 protein expression as well as the ChIP data for CLOCK:BMAL1 and CRY1 over a circadian cycle and its consequences with regard to interactions of core clock proteins with the E-box and the effects of these interactions on transcription of genes (Nr1d1 and Dbp) regulated exclusively by the core TTFL.
Control of DNA Repair by the Circadian Clock
We discovered that the circadian clock controls nucleotide excision repair in mammalian organisms. We wish to apply this finding for cancer prevention and treatment because excision repair removes DNA damage caused by carcinogens as well as damage caused by anticancer drugs. We are using mice as the model organism for these studies. We found that excision repair of carcinogenic UV damage in mice is low at early morning hours and reaches its highest level in the evening. As a consequence, mice exposed to carcinogenic UVB light in morning are 4-times more likely to develop invasive skin cancer than mice exposed to the same UVB dose in the evening (Fig. 3A). These findings should have implications for the optimum time for human sun exposure. To determine the effect of the circadian clock on the repair of DNA damage caused by the anticancer drug cisplatin, we injected cisplatin into mice at 4 hour intervals for a period of one day and analyzed the repair of Platinum-DNA adducts over the course of the day genome-wide and at single nucleotide resolution. We found that the repair of Pt-DNA adducts is controlled by two circadian programs in mouse tissue: (1) The clock controls transcription of 1500-2000 genes with expression maxima of different genes spread over the entire day, but with the majority peaking at pre-dawn and pre-dusk. Because transcription stimulates repair of adducts in the transcribed strand (TS), the TS of circadian-controlled genes are repaired at times of day specific for each gene with prominent peaks at pre-dawn and pre-dusk (Fig. 3B). (2) The clock controls expression of the XPA protein and therefore of excision repair activity which peaks at ZT8-10 (ZT=0 is the time when light is turned on, ZT=12 is the time light is turned off under 12h light: 12h dark conditions). Because the basal repair activity has rhythmicity, the repair of Pt-DNA adducts in the regions of the genome that are not transcribed (the nontranscribed strand (NTS) of transcribed genes, both strands of non-transcribed genes, and intergenic regions) peak at ZT8-10. As a consequence of these two circadian programs, in many circadian-controlled genes the peak and trough repair times for TS and NTS are different, and sometimes anti-phase. Cisplatin is the most commonly used chemotherapeutic drug for treating solid tumors. However, the usefulness of this important drug is limited due of its toxicity and primary acquired resistance by cancer cells. Frequently, biochemical pathways of cancer cells are out of synchrony with those in normal tissues, and these are indications that the circadian clock is “broken” in cancerous tissue. We aim to take advantage of our data regarding orderly repair in normal tissue compared to arrhythmic repair in cancer to develop cisplatin administration regiments (Chronochemotherapy) to improve the efficacy and reduce side effects of cisplatin therapy.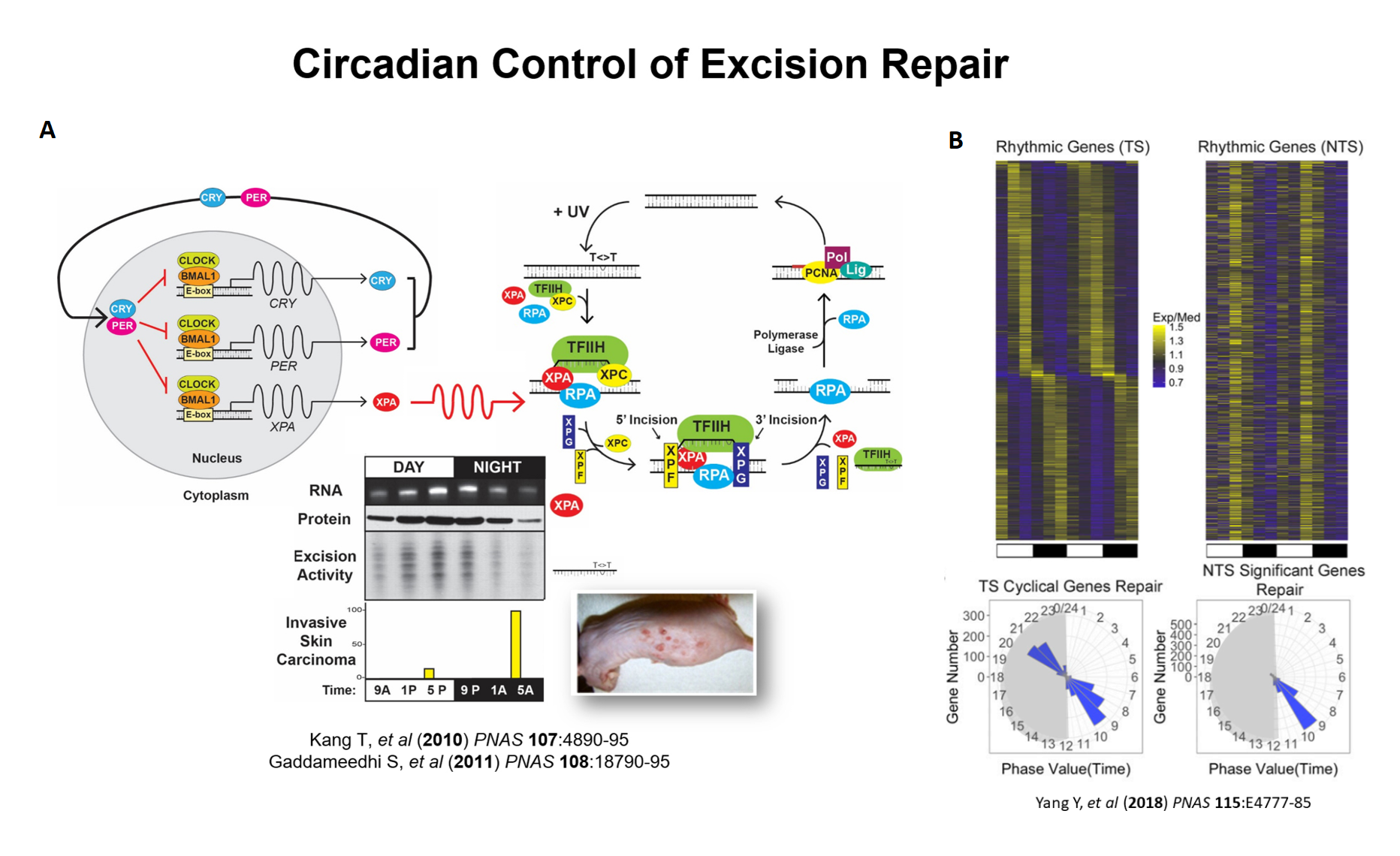
Fig. 3. Control of DNA Repair by the Circadian Clock. (A) Effect of time of day of exposure to UVB on skin carcinogenesis (visual diagnosis) in SKH-1 hairless mice.(B) Heatmaps (top) and radial diagrams (bottom) of circadian repair cycles of the transcribed strand (TS) and the nontranscribed strand (NTS) of 1,661 highly rhythmic genes in mouse kidney.